1 Load packages
library(tidyverse) # data wrangling
library(prada) # function "is_in_tolerance`
library(rstanarm) # Bayes regression
library(easystats) # R2 etc
library(DataExplorer) # data vis
library(tictoc)2 Motivation
Bayes models (using MCMC) build on drawing random numbers. By their very nature, random numbers are random. Unless they are not. As you may know, the random number fuctions in computers are purely deterministic.
However, in practice, some inpredictable behavior may still show up. The reason being simply that two computational environment must - in theory - being exactly identical in order to reproduce the same results. At least identical in every bit that influence random number generation.
Stan results will only be exactly reproducible if all of the following components are identical:
- Stan version
- tan interface (RStan, PyStan, CmdStan) and version, plus version of interface language (R, Python, shell)
- versions of included libraries (Boost and Eigen)
- operating system version
- computer hardware including CPU, motherboard and memory
- C++ compiler, including version, compiler flags, and linked libraries same configuration of call to Stan, including random seed, chain ID, initialization and data
3 But in practice, how large is the difference?
Okay, let’s try to find out the degree of difference to be expected in the process of drawing random numbers.
As a first step, let’s run many regressions and analyze the variance of the results.
Here’s a simple regression with the seed fixed. Please see below for the spec of my machine.
reference_model <- rstanarm::stan_glm(mpg ~ hp,
data = mtcars,
seed = 42,
refresh = 0)
coef(reference_model)
#> (Intercept) hp
#> 30.11668130 -0.06820988And now the same model but without fixing the seed.
out <-
tibble(id = integer(),
b0 = numeric(),
b1 = numeric(),
r2 = numeric()
)
n <- 10^2tic()
for (i in 1:n) {
stan_mod <- rstanarm::stan_glm(mpg ~ hp,
data = mtcars,
refresh = 0)
mod_results <-
tibble(id = i,
b0 = coef(stan_mod)[1],
b1 = coef(stan_mod)[2],
r2 = performance::r2(stan_mod)$R2_Bayes
)
out <- rbind(out, mod_results)
print(glue::glue("i: {i}, b0: {round(coef(stan_mod)[1], 3)}, b1: {round(coef(stan_mod)[2], 3)}, r2: {round(mod_results$r2\n, 3)}"))
}
#> i: 1, b0: 30.083, b1: -0.068, r2: 0.588
#> i: 2, b0: 30.057, b1: -0.068, r2: 0.587
#> i: 3, b0: 30.063, b1: -0.068, r2: 0.585
#> i: 4, b0: 30.038, b1: -0.068, r2: 0.585
#> i: 5, b0: 30.058, b1: -0.068, r2: 0.584
#> i: 6, b0: 30.064, b1: -0.068, r2: 0.585
#> i: 7, b0: 30.108, b1: -0.068, r2: 0.586
#> i: 8, b0: 30.057, b1: -0.068, r2: 0.583
#> i: 9, b0: 30.061, b1: -0.068, r2: 0.588
#> i: 10, b0: 30.097, b1: -0.068, r2: 0.587
#> i: 11, b0: 30.1, b1: -0.068, r2: 0.588
#> i: 12, b0: 30.088, b1: -0.068, r2: 0.583
#> i: 13, b0: 30.083, b1: -0.068, r2: 0.588
#> i: 14, b0: 30.088, b1: -0.068, r2: 0.587
#> i: 15, b0: 30.022, b1: -0.068, r2: 0.584
#> i: 16, b0: 30.085, b1: -0.068, r2: 0.584
#> i: 17, b0: 30.046, b1: -0.068, r2: 0.587
#> i: 18, b0: 30.071, b1: -0.068, r2: 0.584
#> i: 19, b0: 30.136, b1: -0.068, r2: 0.589
#> i: 20, b0: 30.034, b1: -0.068, r2: 0.582
#> i: 21, b0: 30.016, b1: -0.068, r2: 0.588
#> i: 22, b0: 30.073, b1: -0.068, r2: 0.588
#> i: 23, b0: 30.147, b1: -0.068, r2: 0.587
#> i: 24, b0: 30.071, b1: -0.068, r2: 0.588
#> i: 25, b0: 30.077, b1: -0.068, r2: 0.59
#> i: 26, b0: 30.034, b1: -0.068, r2: 0.587
#> i: 27, b0: 30.065, b1: -0.068, r2: 0.585
#> i: 28, b0: 30.061, b1: -0.068, r2: 0.585
#> i: 29, b0: 30.049, b1: -0.068, r2: 0.586
#> i: 30, b0: 30.127, b1: -0.068, r2: 0.586
#> i: 31, b0: 30.078, b1: -0.068, r2: 0.583
#> i: 32, b0: 30.108, b1: -0.068, r2: 0.587
#> i: 33, b0: 30.067, b1: -0.068, r2: 0.586
#> i: 34, b0: 30.086, b1: -0.068, r2: 0.587
#> i: 35, b0: 30.101, b1: -0.068, r2: 0.589
#> i: 36, b0: 30.042, b1: -0.068, r2: 0.588
#> i: 37, b0: 30.024, b1: -0.068, r2: 0.584
#> i: 38, b0: 30.146, b1: -0.068, r2: 0.588
#> i: 39, b0: 30.089, b1: -0.068, r2: 0.585
#> i: 40, b0: 30.147, b1: -0.069, r2: 0.589
#> i: 41, b0: 30.089, b1: -0.068, r2: 0.587
#> i: 42, b0: 30.066, b1: -0.068, r2: 0.585
#> i: 43, b0: 30.064, b1: -0.068, r2: 0.584
#> i: 44, b0: 30.139, b1: -0.069, r2: 0.588
#> i: 45, b0: 30.075, b1: -0.068, r2: 0.589
#> i: 46, b0: 30.065, b1: -0.068, r2: 0.59
#> i: 47, b0: 30.088, b1: -0.068, r2: 0.586
#> i: 48, b0: 30.051, b1: -0.068, r2: 0.586
#> i: 49, b0: 30.116, b1: -0.068, r2: 0.588
#> i: 50, b0: 30.101, b1: -0.068, r2: 0.588
#> i: 51, b0: 30.107, b1: -0.068, r2: 0.584
#> i: 52, b0: 30.093, b1: -0.068, r2: 0.585
#> i: 53, b0: 30.088, b1: -0.068, r2: 0.585
#> i: 54, b0: 30.062, b1: -0.068, r2: 0.587
#> i: 55, b0: 30.068, b1: -0.068, r2: 0.587
#> i: 56, b0: 30.067, b1: -0.068, r2: 0.586
#> i: 57, b0: 30.045, b1: -0.068, r2: 0.586
#> i: 58, b0: 30.038, b1: -0.068, r2: 0.585
#> i: 59, b0: 30.131, b1: -0.068, r2: 0.589
#> i: 60, b0: 30.089, b1: -0.068, r2: 0.588
#> i: 61, b0: 30.075, b1: -0.068, r2: 0.584
#> i: 62, b0: 30.06, b1: -0.068, r2: 0.583
#> i: 63, b0: 30.139, b1: -0.068, r2: 0.59
#> i: 64, b0: 30.074, b1: -0.068, r2: 0.584
#> i: 65, b0: 30.094, b1: -0.068, r2: 0.587
#> i: 66, b0: 30.028, b1: -0.068, r2: 0.587
#> i: 67, b0: 30.102, b1: -0.068, r2: 0.587
#> i: 68, b0: 30.043, b1: -0.068, r2: 0.586
#> i: 69, b0: 30.032, b1: -0.068, r2: 0.583
#> i: 70, b0: 30.088, b1: -0.068, r2: 0.587
#> i: 71, b0: 30.081, b1: -0.068, r2: 0.588
#> i: 72, b0: 30.103, b1: -0.068, r2: 0.589
#> i: 73, b0: 30.098, b1: -0.068, r2: 0.588
#> i: 74, b0: 30.108, b1: -0.068, r2: 0.589
#> i: 75, b0: 30.096, b1: -0.068, r2: 0.586
#> i: 76, b0: 30.104, b1: -0.068, r2: 0.584
#> i: 77, b0: 30.067, b1: -0.068, r2: 0.585
#> i: 78, b0: 30.092, b1: -0.068, r2: 0.588
#> i: 79, b0: 30.086, b1: -0.068, r2: 0.584
#> i: 80, b0: 30.04, b1: -0.068, r2: 0.586
#> i: 81, b0: 30.069, b1: -0.068, r2: 0.587
#> i: 82, b0: 30.128, b1: -0.068, r2: 0.587
#> i: 83, b0: 30.091, b1: -0.068, r2: 0.586
#> i: 84, b0: 30.114, b1: -0.068, r2: 0.586
#> i: 85, b0: 30.084, b1: -0.068, r2: 0.586
#> i: 86, b0: 30.049, b1: -0.068, r2: 0.585
#> i: 87, b0: 30.095, b1: -0.068, r2: 0.586
#> i: 88, b0: 30.012, b1: -0.068, r2: 0.586
#> i: 89, b0: 30.066, b1: -0.068, r2: 0.587
#> i: 90, b0: 30.063, b1: -0.068, r2: 0.587
#> i: 91, b0: 30.034, b1: -0.068, r2: 0.583
#> i: 92, b0: 30.13, b1: -0.068, r2: 0.585
#> i: 93, b0: 30.043, b1: -0.068, r2: 0.585
#> i: 94, b0: 30.033, b1: -0.068, r2: 0.583
#> i: 95, b0: 30.098, b1: -0.068, r2: 0.586
#> i: 96, b0: 30.07, b1: -0.068, r2: 0.586
#> i: 97, b0: 30.082, b1: -0.068, r2: 0.585
#> i: 98, b0: 30.03, b1: -0.068, r2: 0.585
#> i: 99, b0: 30.046, b1: -0.068, r2: 0.585
#> i: 100, b0: 30.051, b1: -0.068, r2: 0.585
toc()
#> 71.804 sec elapsed4 Check if in tolerance region
Let’s define the region of tolerance such that:
tol_rel <- .05
tol_abs <- tol_relCheck for first value manually:
is_in_tolerance(asis = out$b0[1],
tobe = coef(reference_model)[1],
tol_rel = .05,
tol_abs = .05 * sd(mtcars$mpg))
#> [1] TRUEOkay, now check in a loop all values.
4.1 b0
out <-
out |>
mutate(b0_in_tolerance = map_lgl(b0, ~ is_in_tolerance(asis = .x,
tobe = coef(reference_model)[1],
tol_rel = tol_rel,
tol_abs = tol_rel * sd(mtcars$mpg))),
b0_diff_abs = coef(reference_model)[1] - b0,
b0_diff_rel = b0_diff_abs / coef(reference_model)[1])4.2 b1
out <-
out |>
mutate(b1_in_tolerance = map_lgl(b1, ~ is_in_tolerance(asis = .x,
tobe = coef(reference_model)[2],
tol_rel = tol_rel,
tol_abs = tol_rel * sd(mtcars$mpg))),
b1_diff_abs = coef(reference_model)[2] - b1,
b1_diff_rel = b1_diff_abs / coef(reference_model)[2])4.3 R2
out <-
out |>
mutate(r2_in_tolerance = map_lgl(r2, ~ is_in_tolerance(asis = .x,
tobe = r2(reference_model)$R2_Bayes,
tol_rel = tol_rel,
tol_abs = tol_rel)),
r2_diff_abs = r2(reference_model)$R2_Bayes - r2,
r2_diff_rel = r2_diff_abs / coef(reference_model)[2])4.4 Count
count(out, b0_in_tolerance)
#> # A tibble: 1 × 2
#> b0_in_tolerance n
#> <lgl> <int>
#> 1 TRUE 100
count(out, b1_in_tolerance)
#> # A tibble: 1 × 2
#> b1_in_tolerance n
#> <lgl> <int>
#> 1 TRUE 100
count(out, r2_in_tolerance)
#> # A tibble: 1 × 2
#> r2_in_tolerance n
#> <lgl> <int>
#> 1 TRUE 1005 Check variability
out |>
select(-id) |>
describe_distribution(range = TRUE)
#> Variable | Mean | SD | IQR | Range | Skewness | Kurtosis | n | n_Missing
#> -------------------------------------------------------------------------------------------------------------
#> b0 | 30.08 | 0.03 | 0.04 | [30.01, 30.15] | 0.25 | -0.32 | 100 | 0
#> b1 | -0.07 | 1.97e-04 | 2.51e-04 | [-0.07, -0.07] | -0.60 | 0.19 | 100 | 0
#> r2 | 0.59 | 1.81e-03 | 2.86e-03 | [0.58, 0.59] | -0.04 | -0.53 | 100 | 0
#> b0_diff_abs | 0.04 | 0.03 | 0.04 | [-0.03, 0.10] | -0.25 | -0.32 | 100 | 0
#> b0_diff_rel | 1.32e-03 | 1.04e-03 | 1.33e-03 | [-1.02e-03, 3.48e-03] | -0.25 | -0.32 | 100 | 0
#> b1_diff_abs | -1.31e-04 | 1.97e-04 | 2.51e-04 | [-4.79e-04, 4.98e-04] | 0.60 | 0.19 | 100 | 0
#> b1_diff_rel | 1.93e-03 | 2.89e-03 | 3.67e-03 | [-7.31e-03, 7.02e-03] | -0.60 | 0.19 | 100 | 0
#> r2_diff_abs | -2.71e-04 | 1.81e-03 | 2.86e-03 | [-4.53e-03, 3.92e-03] | 0.04 | -0.53 | 100 | 0
#> r2_diff_rel | 3.97e-03 | 0.03 | 0.04 | [-0.06, 0.07] | -0.04 | -0.53 | 100 | 0In comparison, the SD of the y-variable (mpg) is:
describe_distribution(mtcars |> select(mpg))
#> Variable | Mean | SD | IQR | Range | Skewness | Kurtosis | n | n_Missing
#> --------------------------------------------------------------------------------------
#> mpg | 20.09 | 6.03 | 7.53 | [10.40, 33.90] | 0.67 | -0.02 | 32 | 0plot_density(out |> select(-id))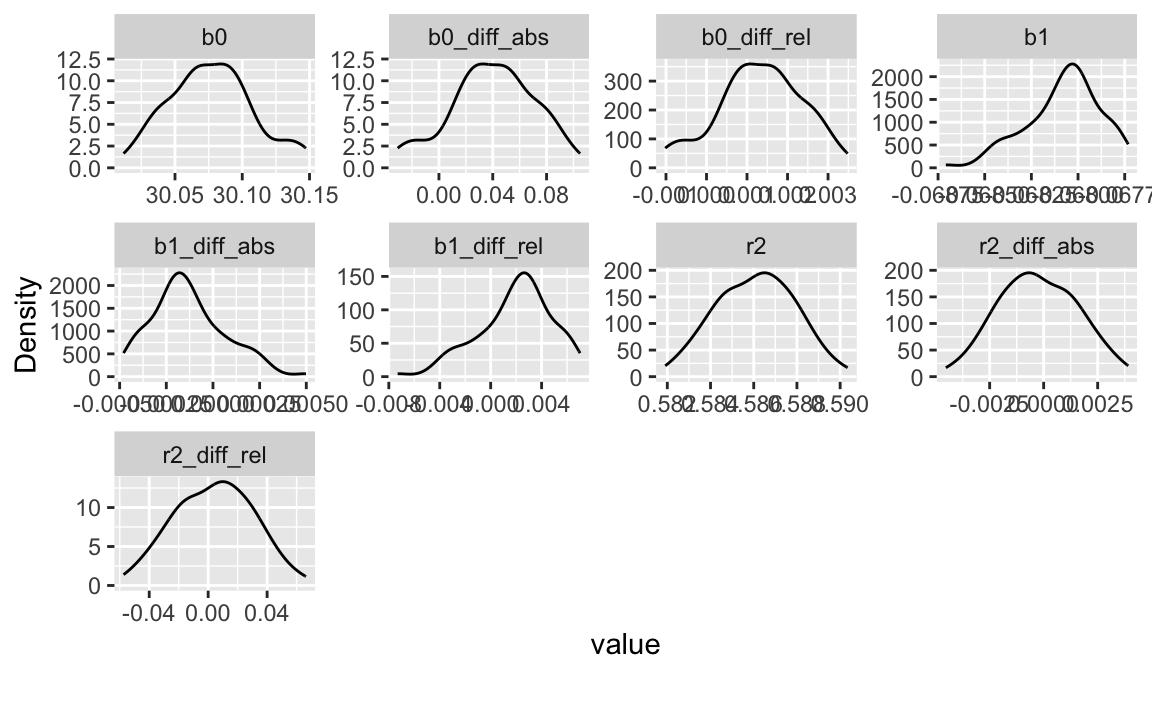
6 Conclusions
The relative deviations of the coefficients from the reference model are in the domaine of . Hence, setting the maximum deviation to should be save.
7 Reproducibility
#> ─ Session info ───────────────────────────────────────────────────────────────────────────────────────────────────────
#> setting value
#> version R version 4.2.1 (2022-06-23)
#> os macOS Big Sur ... 10.16
#> system x86_64, darwin17.0
#> ui X11
#> language (EN)
#> collate en_US.UTF-8
#> ctype en_US.UTF-8
#> tz Europe/Berlin
#> date 2023-12-13
#> pandoc 3.1.1 @ /Applications/RStudio.app/Contents/Resources/app/quarto/bin/tools/ (via rmarkdown)
#>
#> ─ Packages ───────────────────────────────────────────────────────────────────────────────────────────────────────────
#> package * version date (UTC) lib source
#> abind 1.4-5 2016-07-21 [1] CRAN (R 4.2.0)
#> backports 1.4.1 2021-12-13 [1] CRAN (R 4.2.0)
#> base64enc 0.1-3 2015-07-28 [1] CRAN (R 4.2.0)
#> bayesplot 1.10.0 2022-11-16 [1] CRAN (R 4.2.0)
#> bayestestR * 0.13.1 2023-04-07 [1] CRAN (R 4.2.0)
#> blogdown 1.18 2023-06-19 [1] CRAN (R 4.2.0)
#> bookdown 0.36 2023-10-16 [1] CRAN (R 4.2.0)
#> boot 1.3-28.1 2022-11-22 [1] CRAN (R 4.2.0)
#> bslib 0.5.1 2023-08-11 [1] CRAN (R 4.2.0)
#> cachem 1.0.8 2023-05-01 [1] CRAN (R 4.2.0)
#> callr 3.7.3 2022-11-02 [1] CRAN (R 4.2.0)
#> checkmate 2.2.0 2023-04-27 [1] CRAN (R 4.2.0)
#> cli 3.6.1 2023-03-23 [1] CRAN (R 4.2.0)
#> coda 0.19-4 2020-09-30 [1] CRAN (R 4.2.0)
#> codetools 0.2-19 2023-02-01 [1] CRAN (R 4.2.0)
#> colorout * 1.3-0 2023-11-08 [1] Github (jalvesaq/colorout@8384882)
#> colorspace 2.1-0 2023-01-23 [1] CRAN (R 4.2.0)
#> colourpicker 1.3.0 2023-08-21 [1] CRAN (R 4.2.0)
#> correlation * 0.8.4 2023-04-06 [1] CRAN (R 4.2.1)
#> crayon 1.5.2 2022-09-29 [1] CRAN (R 4.2.1)
#> crosstalk 1.2.1 2023-11-23 [1] CRAN (R 4.2.1)
#> curl 5.1.0 2023-10-02 [1] CRAN (R 4.2.0)
#> data.table 1.14.8 2023-02-17 [1] CRAN (R 4.2.0)
#> DataExplorer * 0.8.2 2020-12-15 [1] CRAN (R 4.2.0)
#> datawizard * 0.9.0 2023-09-15 [1] CRAN (R 4.2.0)
#> devtools 2.4.5 2022-10-11 [1] CRAN (R 4.2.1)
#> digest 0.6.33 2023-07-07 [1] CRAN (R 4.2.0)
#> distributional 0.3.2 2023-03-22 [1] CRAN (R 4.2.0)
#> dplyr * 1.1.3 2023-09-03 [1] CRAN (R 4.2.0)
#> DT 0.30 2023-10-05 [1] CRAN (R 4.2.0)
#> dygraphs 1.1.1.6 2018-07-11 [1] CRAN (R 4.2.0)
#> easystats * 0.7.0 2023-11-05 [1] CRAN (R 4.2.1)
#> effectsize * 0.8.6 2023-09-14 [1] CRAN (R 4.2.0)
#> ellipsis 0.3.2 2021-04-29 [1] CRAN (R 4.2.0)
#> emmeans 1.8.9 2023-10-17 [1] CRAN (R 4.2.0)
#> estimability 1.4.1 2022-08-05 [1] CRAN (R 4.2.0)
#> evaluate 0.21 2023-05-05 [1] CRAN (R 4.2.0)
#> fansi 1.0.5 2023-10-08 [1] CRAN (R 4.2.0)
#> farver 2.1.1 2022-07-06 [1] CRAN (R 4.2.0)
#> fastmap 1.1.1 2023-02-24 [1] CRAN (R 4.2.0)
#> forcats * 1.0.0 2023-01-29 [1] CRAN (R 4.2.0)
#> fs 1.6.3 2023-07-20 [1] CRAN (R 4.2.0)
#> generics 0.1.3 2022-07-05 [1] CRAN (R 4.2.0)
#> ggplot2 * 3.4.4 2023-10-12 [1] CRAN (R 4.2.0)
#> glue 1.6.2 2022-02-24 [1] CRAN (R 4.2.0)
#> gridExtra 2.3 2017-09-09 [1] CRAN (R 4.2.0)
#> gtable 0.3.4 2023-08-21 [1] CRAN (R 4.2.0)
#> gtools 3.9.4 2022-11-27 [1] CRAN (R 4.2.0)
#> highr 0.10 2022-12-22 [1] CRAN (R 4.2.0)
#> hms 1.1.3 2023-03-21 [1] CRAN (R 4.2.0)
#> htmltools 0.5.6.1 2023-10-06 [1] CRAN (R 4.2.0)
#> htmlwidgets 1.6.2 2023-03-17 [1] CRAN (R 4.2.0)
#> httpuv 1.6.11 2023-05-11 [1] CRAN (R 4.2.0)
#> igraph 1.5.1 2023-08-10 [1] CRAN (R 4.2.0)
#> inline 0.3.19 2021-05-31 [1] CRAN (R 4.2.0)
#> insight * 0.19.7 2023-11-26 [1] CRAN (R 4.2.1)
#> jquerylib 0.1.4 2021-04-26 [1] CRAN (R 4.2.0)
#> jsonlite 1.8.7 2023-06-29 [1] CRAN (R 4.2.0)
#> knitr 1.45 2023-10-30 [1] CRAN (R 4.2.1)
#> labeling 0.4.3 2023-08-29 [1] CRAN (R 4.2.0)
#> later 1.3.1 2023-05-02 [1] CRAN (R 4.2.0)
#> lattice 0.21-8 2023-04-05 [1] CRAN (R 4.2.0)
#> lifecycle 1.0.4 2023-11-07 [1] CRAN (R 4.2.1)
#> lme4 1.1-34 2023-07-04 [1] CRAN (R 4.2.0)
#> loo 2.6.0 2023-03-31 [1] CRAN (R 4.2.0)
#> lubridate * 1.9.3 2023-09-27 [1] CRAN (R 4.2.0)
#> magrittr 2.0.3 2022-03-30 [1] CRAN (R 4.2.0)
#> markdown 1.11 2023-10-19 [1] CRAN (R 4.2.0)
#> MASS 7.3-60 2023-05-04 [1] CRAN (R 4.2.0)
#> Matrix 1.5-4.1 2023-05-18 [1] CRAN (R 4.2.0)
#> matrixStats 0.63.0 2022-11-18 [1] CRAN (R 4.2.0)
#> memoise 2.0.1 2021-11-26 [1] CRAN (R 4.2.0)
#> mime 0.12 2021-09-28 [1] CRAN (R 4.2.0)
#> miniUI 0.1.1.1 2018-05-18 [1] CRAN (R 4.2.0)
#> minqa 1.2.5 2022-10-19 [1] CRAN (R 4.2.1)
#> modelbased * 0.8.6 2023-01-13 [1] CRAN (R 4.2.1)
#> multcomp 1.4-25 2023-06-20 [1] CRAN (R 4.2.0)
#> munsell 0.5.0 2018-06-12 [1] CRAN (R 4.2.0)
#> mvtnorm 1.2-2 2023-06-08 [1] CRAN (R 4.2.0)
#> networkD3 0.4 2017-03-18 [1] CRAN (R 4.2.0)
#> nlme 3.1-162 2023-01-31 [1] CRAN (R 4.2.0)
#> nloptr 2.0.3 2022-05-26 [1] CRAN (R 4.2.0)
#> parameters * 0.21.3 2023-11-02 [1] CRAN (R 4.2.1)
#> performance * 0.10.8 2023-10-30 [1] CRAN (R 4.2.1)
#> pillar 1.9.0 2023-03-22 [1] CRAN (R 4.2.0)
#> pkgbuild 1.4.0 2022-11-27 [1] CRAN (R 4.2.0)
#> pkgconfig 2.0.3 2019-09-22 [1] CRAN (R 4.2.0)
#> pkgload 1.3.2.1 2023-07-08 [1] CRAN (R 4.2.0)
#> plyr 1.8.8 2022-11-11 [1] CRAN (R 4.2.0)
#> png 0.1-8 2022-11-29 [1] CRAN (R 4.2.0)
#> posterior 1.5.0 2023-10-31 [1] CRAN (R 4.2.1)
#> prada * 0.2.2 2023-12-08 [1] local
#> prettyunits 1.1.1 2020-01-24 [1] CRAN (R 4.2.0)
#> processx 3.8.2 2023-06-30 [1] CRAN (R 4.2.0)
#> profvis 0.3.8 2023-05-02 [1] CRAN (R 4.2.0)
#> promises 1.2.1 2023-08-10 [1] CRAN (R 4.2.0)
#> ps 1.7.5 2023-04-18 [1] CRAN (R 4.2.0)
#> purrr * 1.0.2 2023-08-10 [1] CRAN (R 4.2.0)
#> QuickJSR 1.0.5 2023-09-03 [1] CRAN (R 4.2.0)
#> R6 2.5.1 2021-08-19 [1] CRAN (R 4.2.0)
#> Rcpp * 1.0.11 2023-07-06 [1] CRAN (R 4.2.0)
#> RcppParallel 5.1.7 2023-02-27 [1] CRAN (R 4.2.0)
#> readr * 2.1.4 2023-02-10 [1] CRAN (R 4.2.0)
#> remotes 2.4.2.1 2023-07-18 [1] CRAN (R 4.2.0)
#> report * 0.5.8 2023-12-07 [1] CRAN (R 4.2.1)
#> reshape2 1.4.4 2020-04-09 [1] CRAN (R 4.2.0)
#> rlang 1.1.1 2023-04-28 [1] CRAN (R 4.2.0)
#> rmarkdown 2.25 2023-09-18 [1] CRAN (R 4.2.0)
#> rstan 2.32.3 2023-10-15 [1] CRAN (R 4.2.0)
#> rstanarm * 2.26.1 2023-09-13 [1] CRAN (R 4.2.0)
#> rstantools 2.3.1.1 2023-07-18 [1] CRAN (R 4.2.0)
#> rstudioapi 0.15.0 2023-07-07 [1] CRAN (R 4.2.0)
#> sandwich 3.0-2 2022-06-15 [1] CRAN (R 4.2.0)
#> sass 0.4.7 2023-07-15 [1] CRAN (R 4.2.0)
#> scales 1.2.1 2022-08-20 [1] CRAN (R 4.2.0)
#> see * 0.8.1 2023-11-03 [1] CRAN (R 4.2.1)
#> sessioninfo 1.2.2 2021-12-06 [1] CRAN (R 4.2.0)
#> shiny 1.8.0 2023-11-17 [1] CRAN (R 4.2.1)
#> shinyjs 2.1.0 2021-12-23 [1] CRAN (R 4.2.0)
#> shinystan 2.6.0 2022-03-03 [1] CRAN (R 4.2.0)
#> shinythemes 1.2.0 2021-01-25 [1] CRAN (R 4.2.0)
#> StanHeaders 2.26.28 2023-09-07 [1] CRAN (R 4.2.0)
#> stringi 1.7.12 2023-01-11 [1] CRAN (R 4.2.0)
#> stringr * 1.5.1 2023-11-14 [1] CRAN (R 4.2.1)
#> survival 3.5-5 2023-03-12 [1] CRAN (R 4.2.0)
#> tensorA 0.36.2 2020-11-19 [1] CRAN (R 4.2.0)
#> TH.data 1.1-2 2023-04-17 [1] CRAN (R 4.2.0)
#> threejs 0.3.3 2020-01-21 [1] CRAN (R 4.2.0)
#> tibble * 3.2.1 2023-03-20 [1] CRAN (R 4.2.0)
#> tictoc * 1.2 2023-04-23 [1] CRAN (R 4.2.0)
#> tidyr * 1.3.0 2023-01-24 [1] CRAN (R 4.2.0)
#> tidyselect 1.2.0 2022-10-10 [1] CRAN (R 4.2.0)
#> tidyverse * 2.0.0 2023-02-22 [1] CRAN (R 4.2.0)
#> timechange 0.2.0 2023-01-11 [1] CRAN (R 4.2.0)
#> tzdb 0.4.0 2023-05-12 [1] CRAN (R 4.2.0)
#> urlchecker 1.0.1 2021-11-30 [1] CRAN (R 4.2.0)
#> usethis 2.2.2 2023-07-06 [1] CRAN (R 4.2.0)
#> utf8 1.2.3 2023-01-31 [1] CRAN (R 4.2.0)
#> V8 4.4.0 2023-10-09 [1] CRAN (R 4.2.0)
#> vctrs 0.6.4 2023-10-12 [1] CRAN (R 4.2.0)
#> withr 2.5.2 2023-10-30 [1] CRAN (R 4.2.1)
#> xfun 0.40 2023-08-09 [1] CRAN (R 4.2.0)
#> xtable 1.8-4 2019-04-21 [1] CRAN (R 4.2.0)
#> xts 0.13.1 2023-04-16 [1] CRAN (R 4.2.0)
#> yaml 2.3.7 2023-01-23 [1] CRAN (R 4.2.0)
#> zoo 1.8-12 2023-04-13 [1] CRAN (R 4.2.0)
#>
#> [1] /Users/sebastiansaueruser/Rlibs
#> [2] /Library/Frameworks/R.framework/Versions/4.2/Resources/library
#>
#> ──────────────────────────────────────────────────────────────────────────────────────────────────────────────────────