Load packages
library(tidyverse) # data wrangling
library(mosaic) # funplot
library(plotly) # interactive plots
library(scatterplot3d) # nomen est omen
library(rsm) # 3d scatterplotsMotivation
Linear models are a standard way of predicting or explaining some data. Visualizing data is not only of didactical value but provides heuristical value too, as demonstrated by Anscombe’s Quartet.
Visualizing linear models in 2D is straightforward, but visualizing linear models with more than one predictor is much less so. The aim of this post is to demonstrate some ways do visualize linear models with more than one predictor, using popular R packages. We will focus on 3D examples, that is, two predictors.
Setup
Some data
data(mtcars)Some packages
library(tidyverse)
library(mosaic)Baseline model
For the sake of simplicity, let’s start with a 2D model.
lm0 <- lm(mpg ~ hp, data = mtcars)`mosaic``
plotModel(lm0)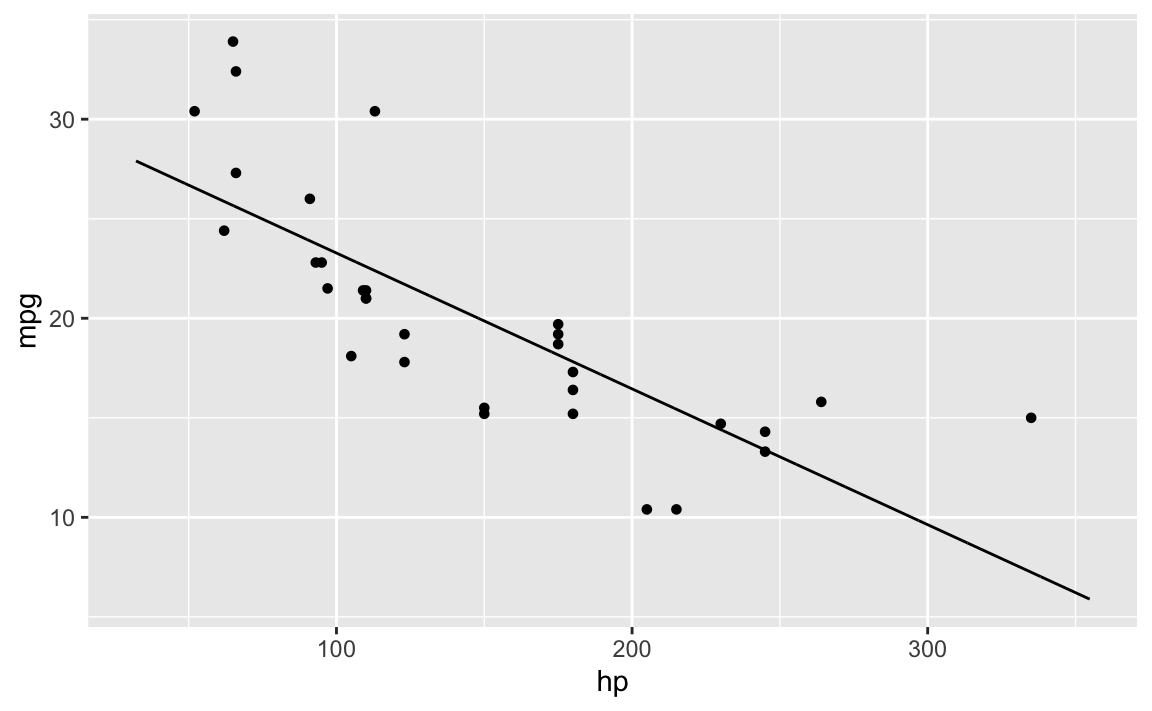
`ggformula``
gf_point(mpg ~ hp, data = mtcars) %>%
gf_lm()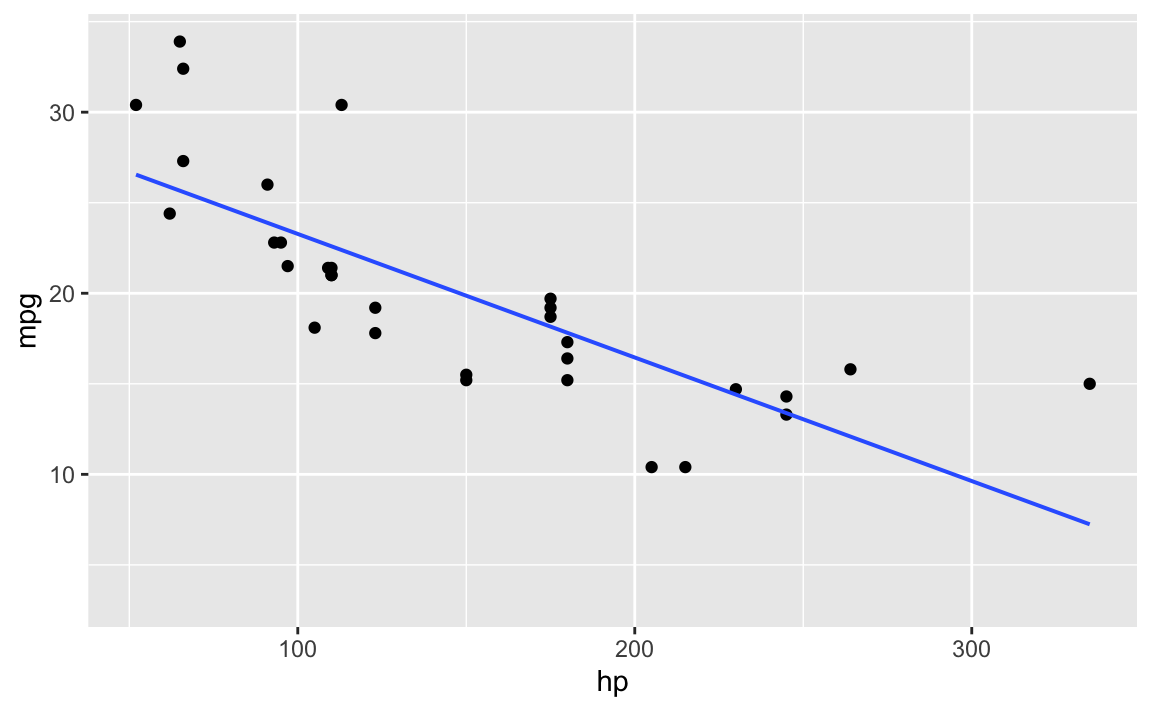
ggplot2
f0 <- function(x) {coef(lm0)[1] + coef(lm0)[2] * x}
ggplot(mtcars) +
aes(x = hp, y = mpg) +
geom_point() +
stat_function(fun = f0)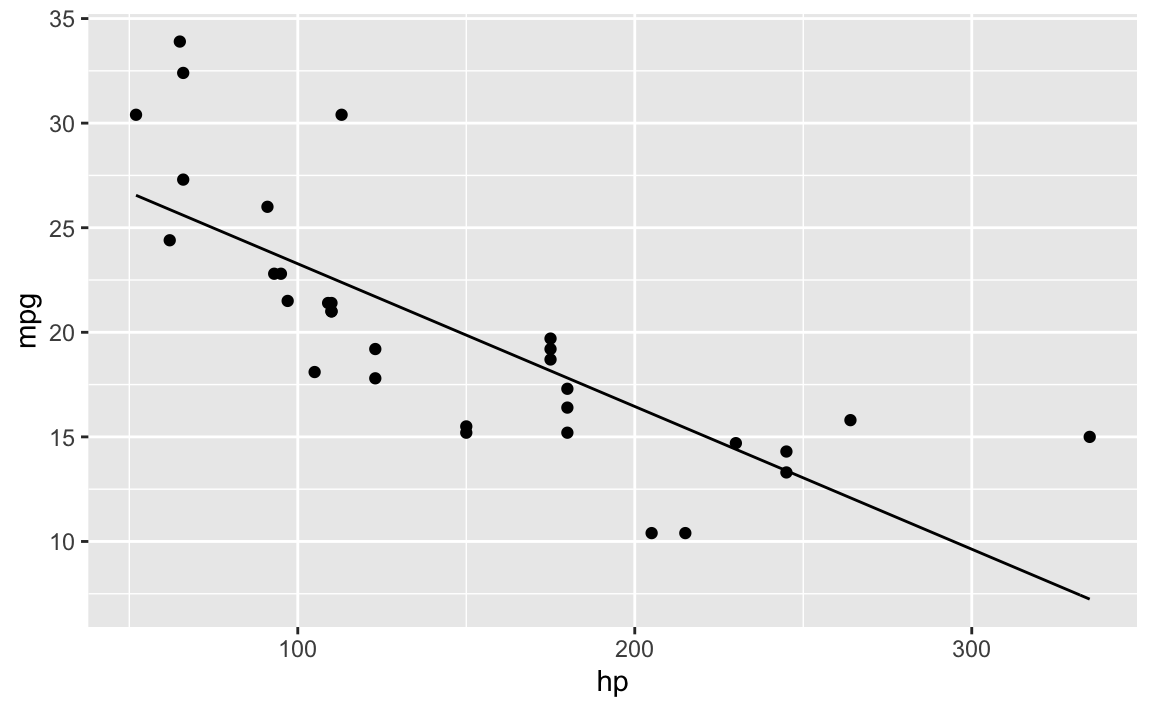
makeFun
Extracting the function itself from a linear model can be achieved like this:
f0 <- makeFun(lm0)
f0
#> function (hp, ..., transformation = function (x)
#> x)
#> return(transformation(predict(model, newdata = data.frame(hp = hp),
#> ...)))
#> <environment: 0x7f8528a74eb0>
#> attr(,"coefficients")
#> (Intercept) hp
#> 30.09886054 -0.06822828Then again:
ggplot(mtcars) +
aes(x = hp, y = mpg) +
geom_point() +
stat_function(fun = f0)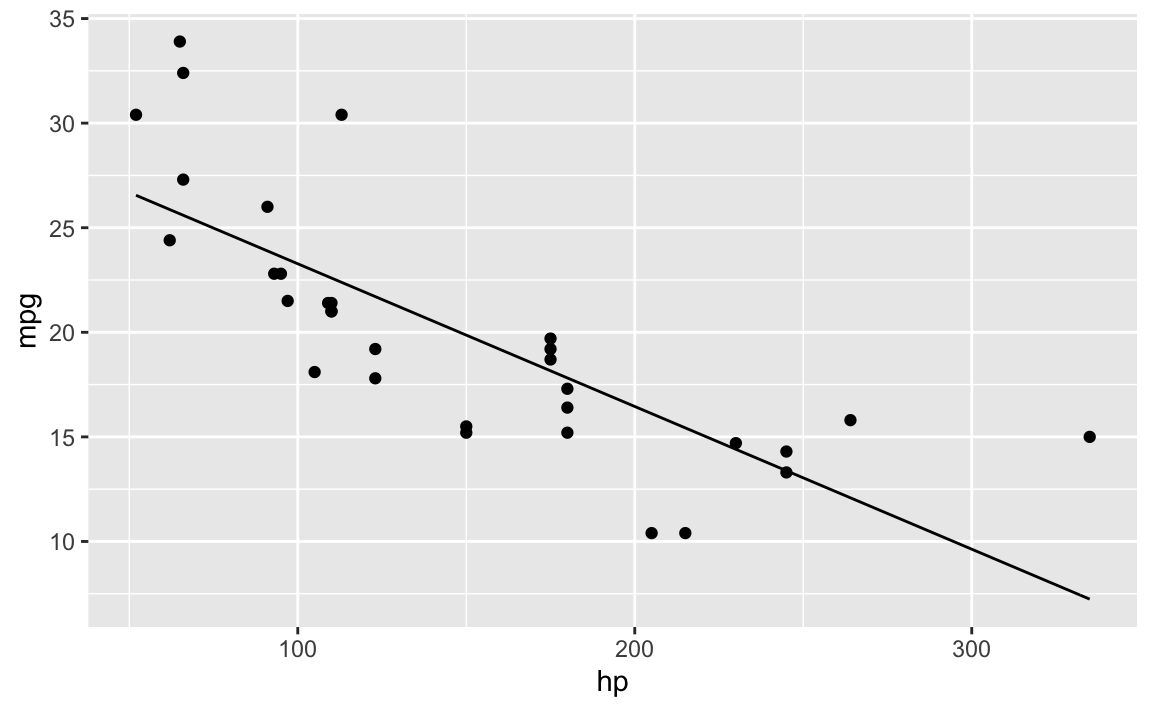
Our (initial) linear model for 3D
lm1 <- lm(mpg ~ hp * disp, data = mtcars)Note that this notation is synonymous to this one:
lm1 <- lm(mpg ~ hp + disp + hp:disp, data = mtcars)gf_function_2d()
Let’s briefly check the range of the predictors:
preds <- c("hp", "disp")mtcars %>%
dplyr::select(one_of(preds)) %>%
summarise_all(funs(min, max))
#> hp_min disp_min hp_max disp_max
#> 1 52 71.1 335 472We’ll use this information to gauge the limits of the following graph:
f1 <- makeFun(lm1)
p1 <- gf_function_2d(f1, xlim = c(50, 300), ylim = c(50, 500)) %>%
gf_labs(x = "hp", y = "disp") %>%
gf_refine(scale_fill_viridis_c())
p1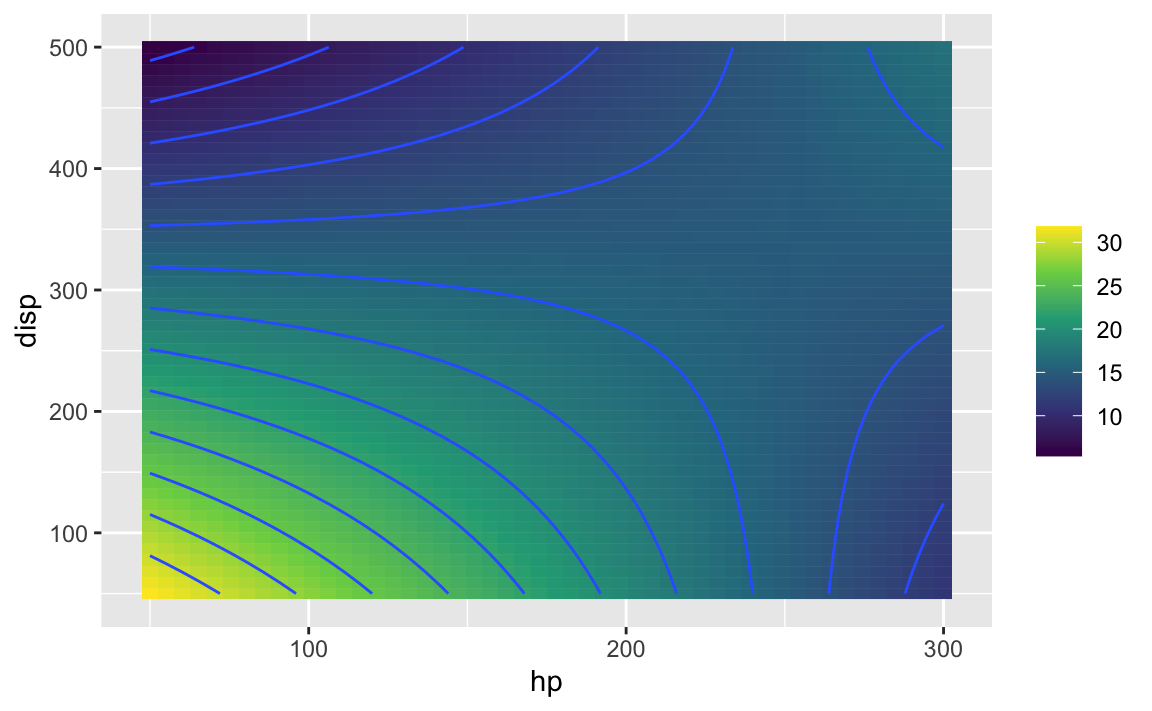
Computing a grid and plotting it
First we devise seom combinations of hp and disp values. Then we predict the mpg values based on those values using lm1.
hp_disp_grid <- expand_grid(hp = seq(50, 300, by = 10), disp = seq(50, 500, by= 10))
grid2 <- hp_disp_grid %>%
mutate(mpg_pred_lm1 = predict(lm1, newdata = data.frame(hp, disp)))Check
gf_density(~mpg_pred_lm1, data = grid2)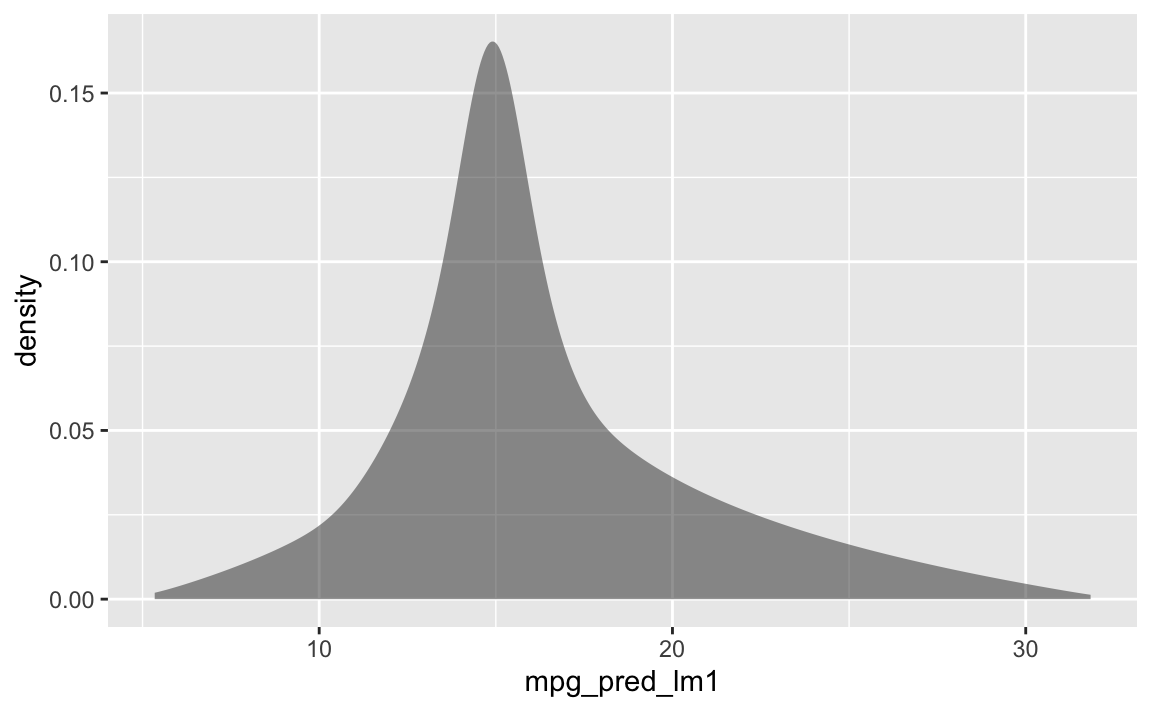
Now add the predicted value to the mtcars dataframe too:
mtcars <- mtcars %>%
mutate(pred_lm1 = predict(lm1))ggplot(aes(x = hp, y = disp), data = grid2) +
geom_raster(aes(fill = mpg_pred_lm1)) +
scale_fill_viridis_c() +
geom_point(data = mtcars, aes(color = pred_lm1)) +
scale_color_viridis_c()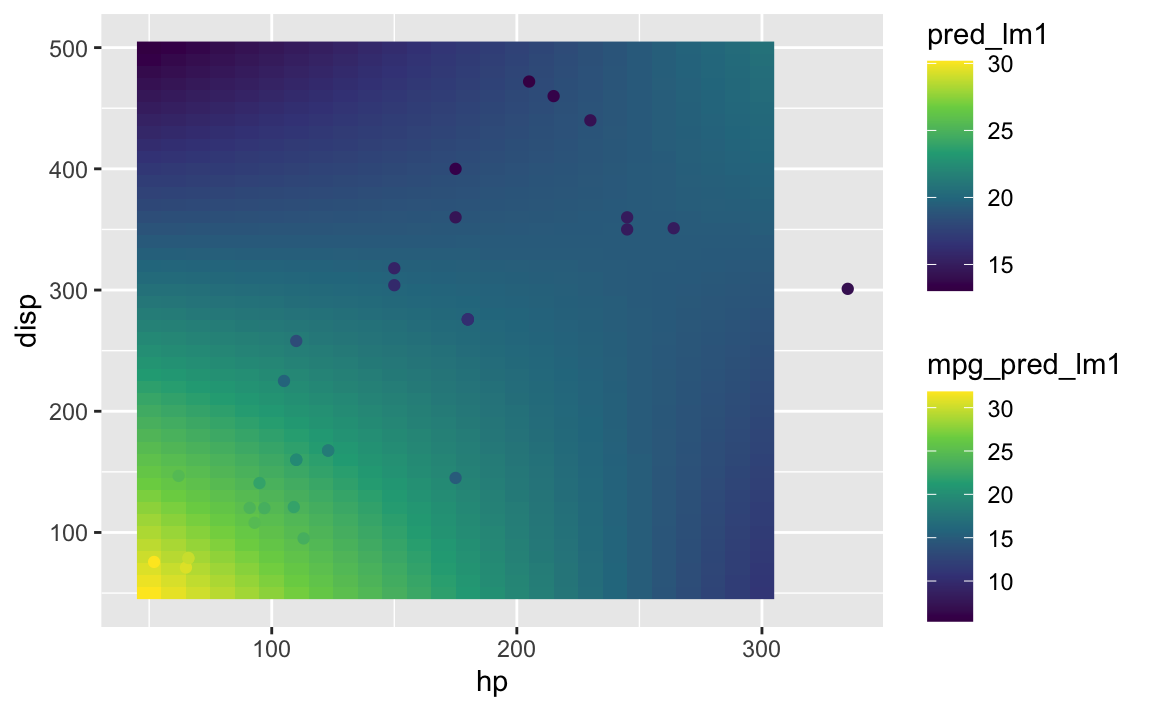
plotFun as tile plot (heatmap)
mosaic provides a convenient function to plot linear models with predictors, plotFun().
plotFun(f1(hp, disp) ~ hp & disp)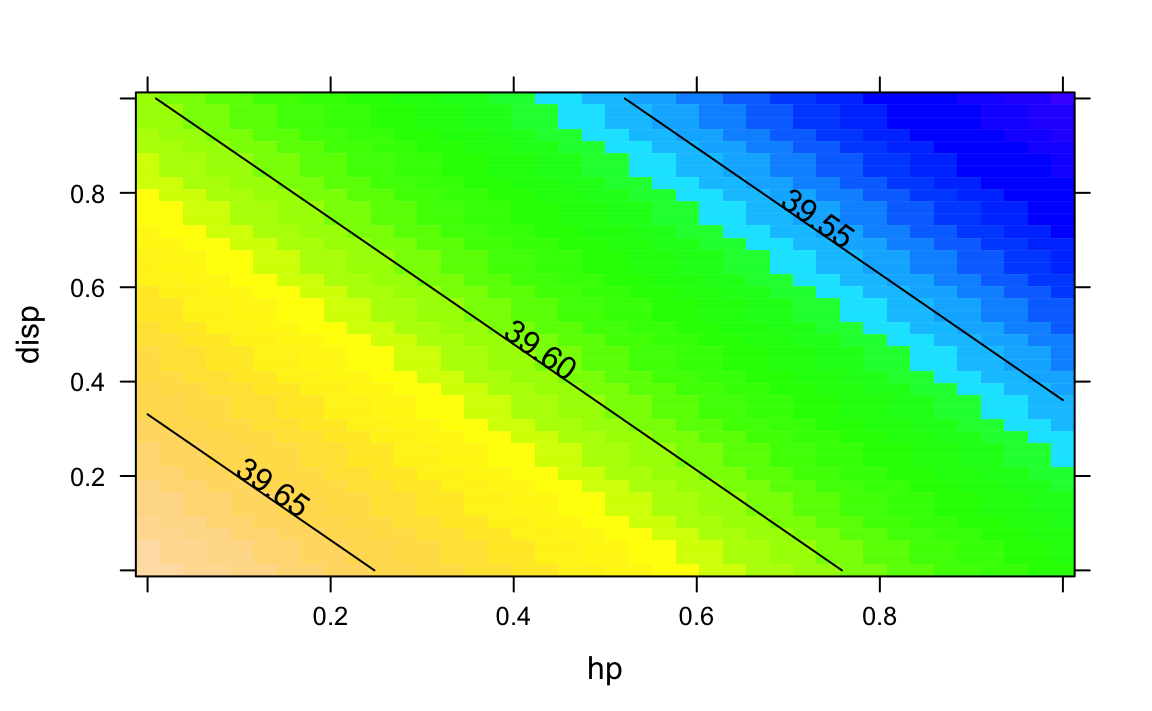
plotFun as 3d surface plot
plotFun(f1(hp, disp) ~ hp & disp, surface = TRUE)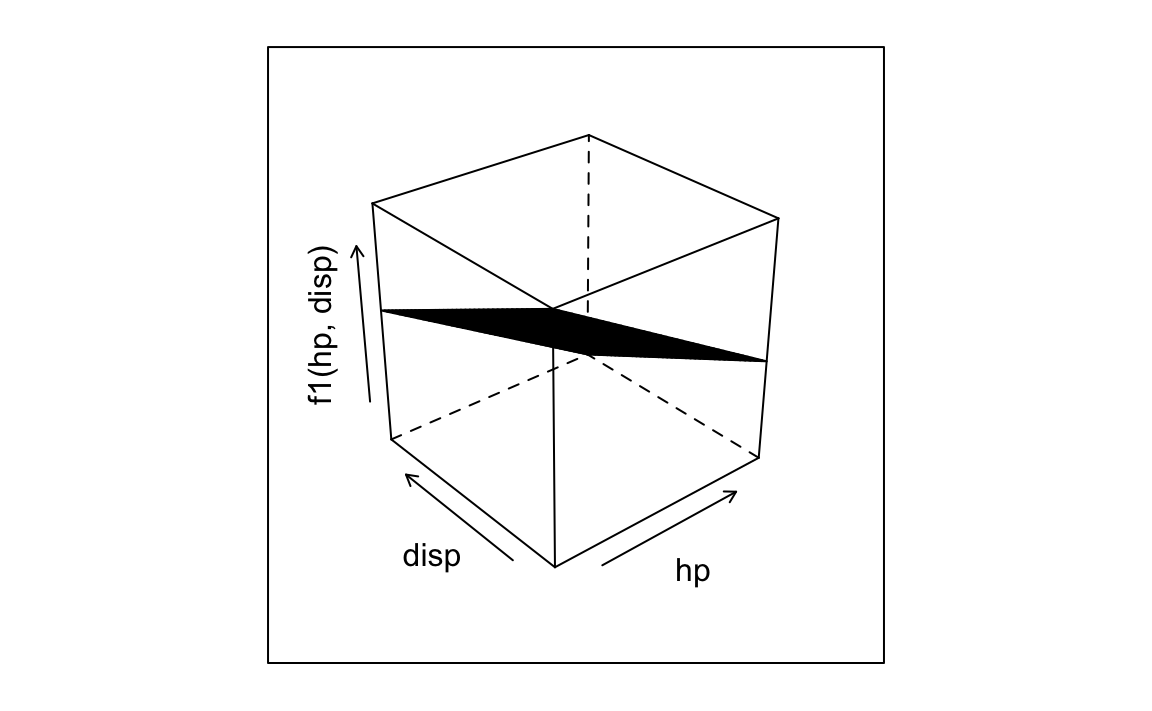
Plotly surface
For plotly surface, we need the data in wide format, ie., we need to spread them out, from long (tidy, standard) format to the wide format. In addition, there should be no more name (key) column, and it ought to be a matrix:
grid_wide <- grid2 %>%
pivot_wider(names_from = disp, values_from = mpg_pred_lm1) %>%
select(-1) %>%
as.matrix
rownames(grid_wide) <- seq(50, 300, by = 10)xaxis <- list(
title = "hp"
#range = c(50, 300)
)
yaxis <- list(
title = "disp"
#range = c(50, 500)
)
zaxis <- list(
title = "mpg"
#range = c(10, 30)
)
p <- plot_ly(z = ~grid_wide,
x = seq(50, 300, by = 10),
y = seq(50, 500, by = 10)) %>%
add_surface(
contours = list(
z = list(
show=TRUE,
usecolormap=TRUE,
highlightcolor="#ff0000"
#project=list(z=TRUE)
)
)
) %>%
layout(
title = "lm1 as 3d plane",
scene = list(
xaxis = xaxis,
yaxis = yaxis,
zaxis = zaxis
))
ppersp()
x <- seq(min(mtcars$hp), max(mtcars$hp), length.out = 25)
y <- seq(min(mtcars$disp), max(mtcars$disp), length.out = 25)
z <- outer(x, y, function(x,y) {predict(lm1, data.frame(hp = x, disp = y))})
persp(x,y,z , theta = c(45, 135, 225, 315), col = "lightblue")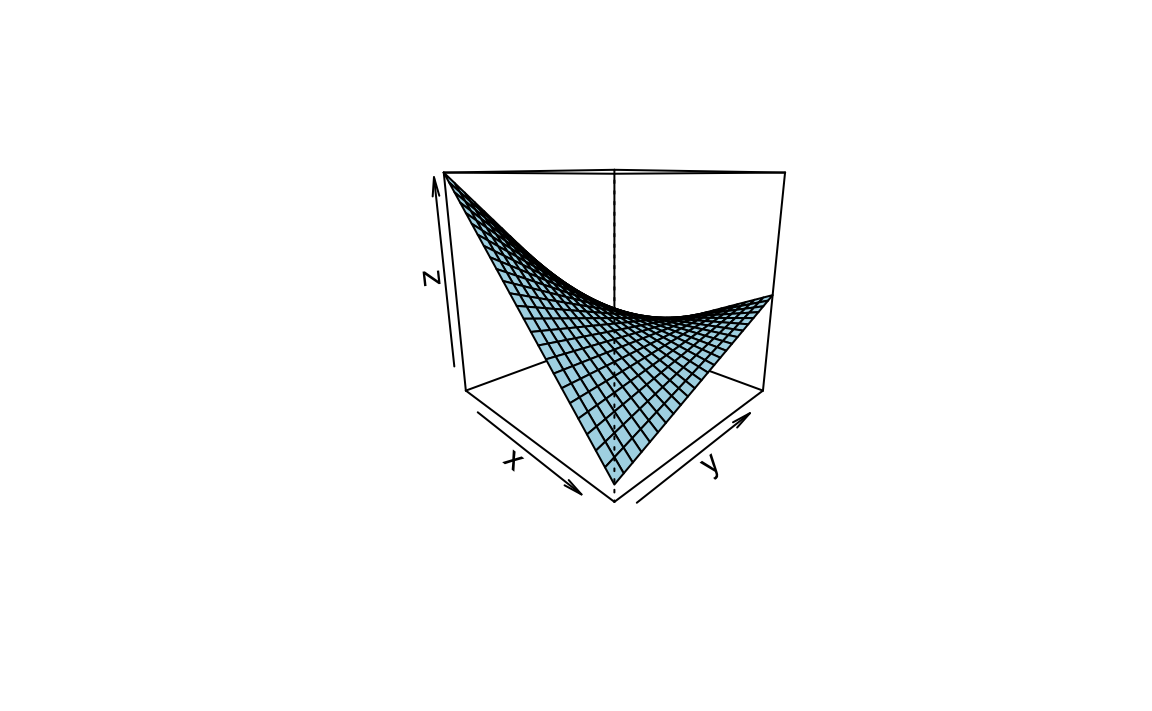
contour()
From package rsm we can make use of contour():
contour(lm1, hp ~ disp, image = TRUE)
contour(lm1, hp ~ disp, image = FALSE)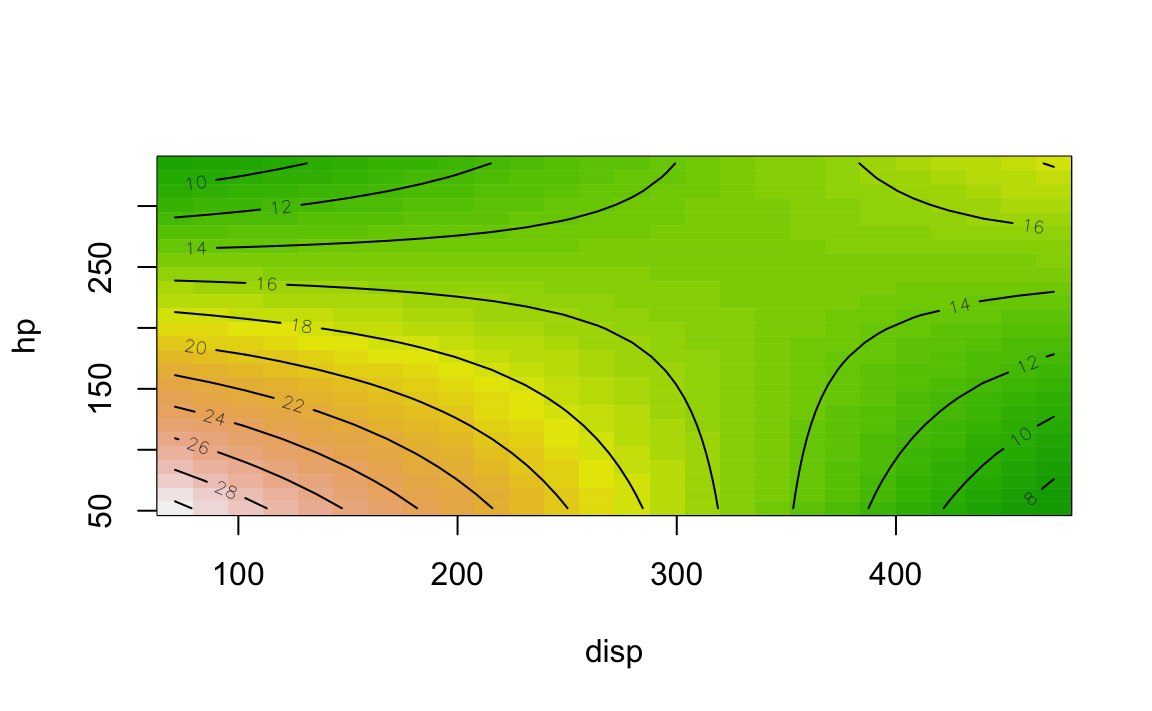
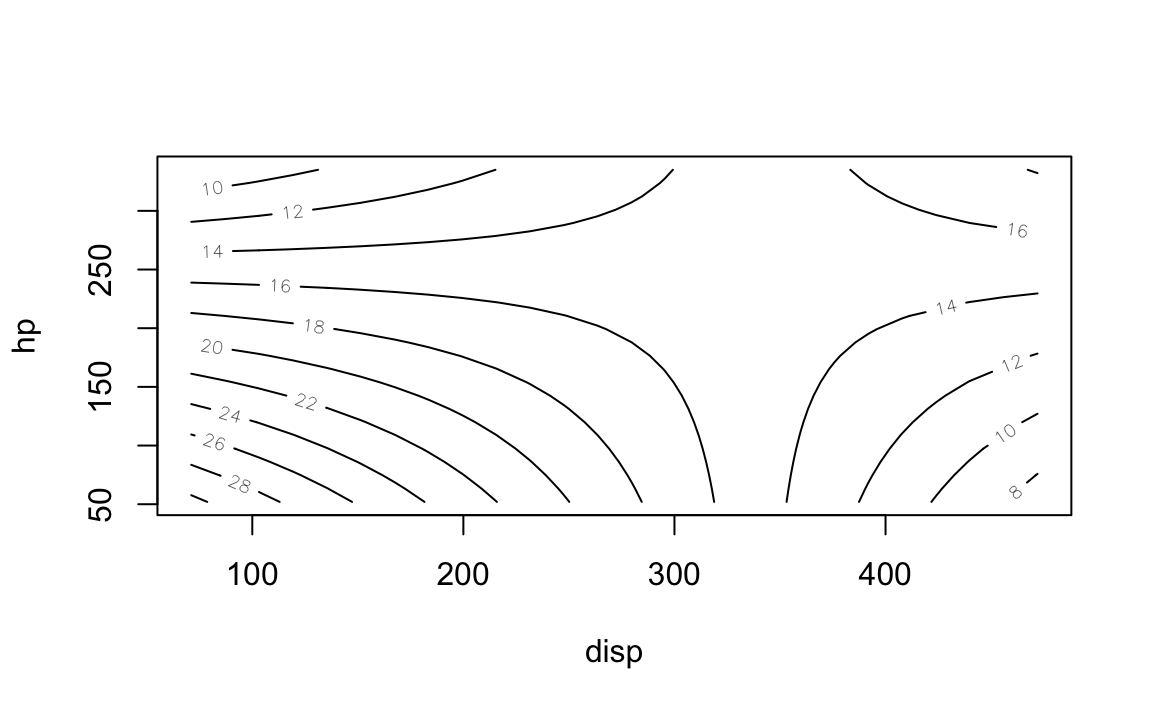
Scatterplot 3d
First plot the scatterplot:
d <- mtcars %>%
select(hp, disp, mpg)
lm1a <- lm(d$mpg ~ d$hp + d$disp)
p3 <- scatterplot3d(d)
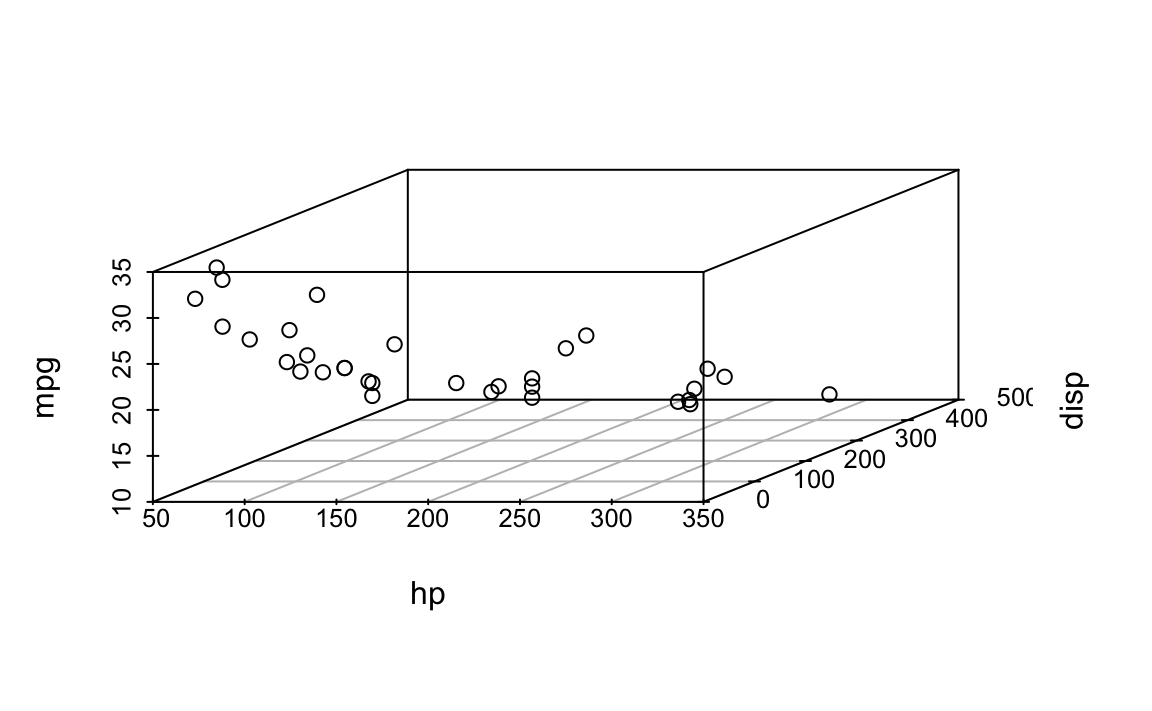
Now add the regression plane:
p3$plane3d(lm1a)
#> Error in segments(x, z1, x + y.max * yx.f, z2 + yz.f * y.max, lty = ltya, : plot.new has not been called yetFor some reason, this code does not run in (my) rmarkdown setup, but it worked (on my machine) in a ordinary R script.